| RESEARCH | MEDIA/NEWS | PUBLICATIONS | SOFTWARE | PEOPLE | ACTIVITIES | ADDRESS | BioImage Informatics |

| Nature, Vol. 598's Cover Story of the BICCN and Whole Brain Full Neuron Morphometry. |

| Current Biology, Vol. 30's Cover Story of the Demselfly and Dragonfly project. More news coverage of this project appeared in New York Times. |
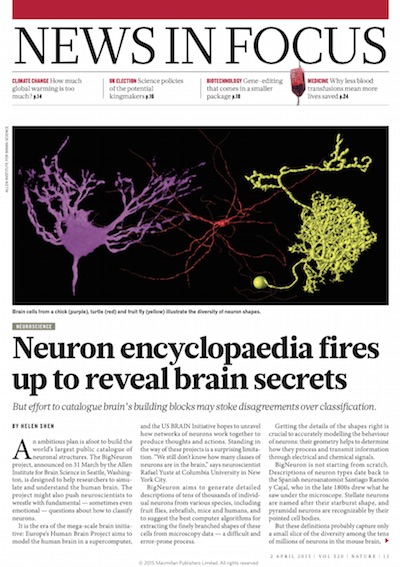
| Nature's 2015 News Highlight of the BigNeuron project. More news coverage of this project appeared in Science, NPR, NBC, etc. |
| Virtual Finger technique published in Nature Communications in July 2014. See the Allen Institute Press Release. |

| A mouse brain wiring map generated at Allen Institute for Brain Science, and published in Nature in April 2014. |

| A SmartScope microimaging system being built in Peng Lab. This picture was used as the cover art of the 2012 Turning Images to Knowledge conference, Oct 28-31, 2012. |
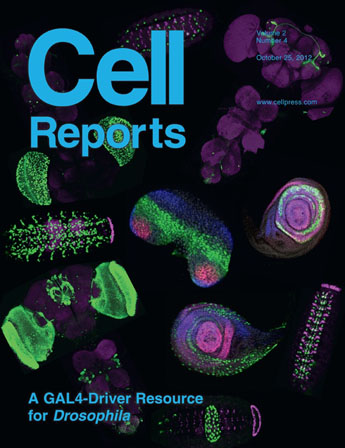
| A collaboration project of the GAL4 resource with G. Rubin lab. My BrainAligner software (Nature Methods, 2011) is used as a critical element for this study, which is published as cover story of Cell Reports 2012. |

| High-throughput mapping of fruit fly brain circuit based on analyzing tens of thousands of 3D laser scanning microscopy image stacks of the adult Drosophila brain. This picture was used as the cover art of the 2011 Bioimage Informatics conference, Sept 18-21, 2011. Particularly, this work is based on a new 3D image registration method called BrainAligner (Nature Methods, 2011). |
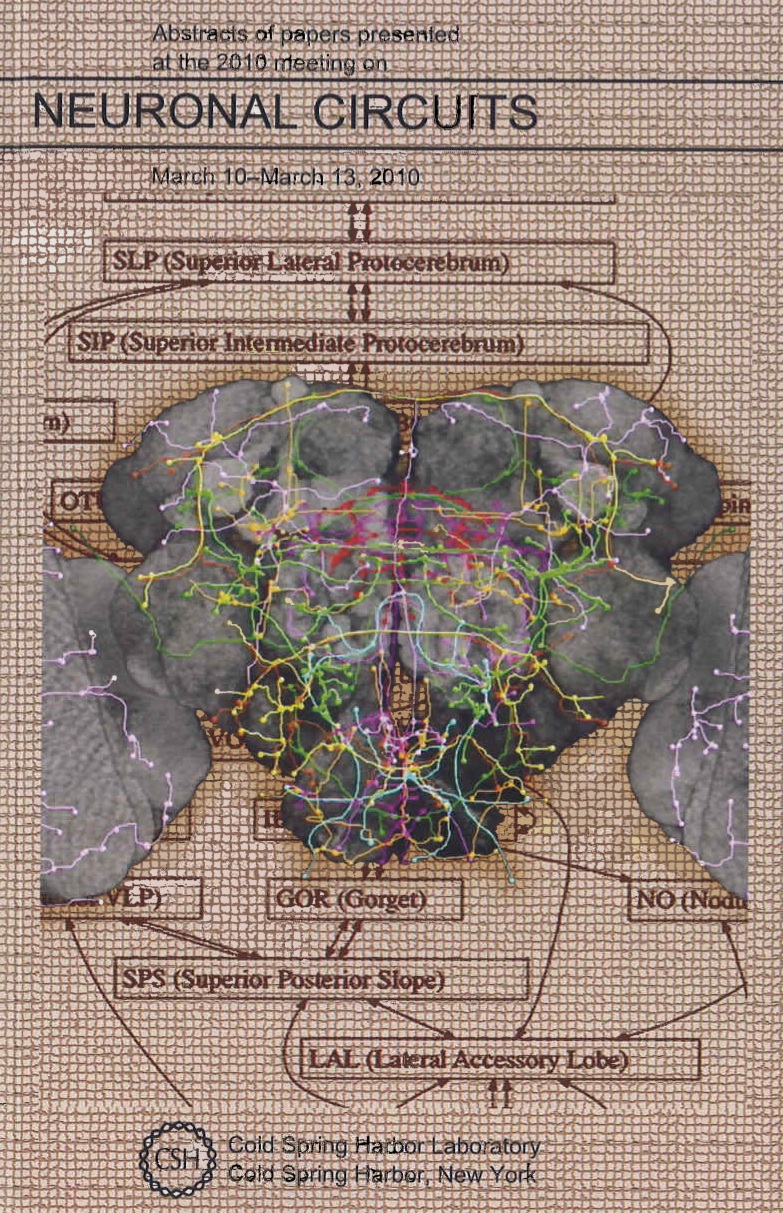
| A neuronal wiring atlas across brain compartments throughout a fruit fly's brain. I developed a pipeline (3D image registration/alignment, neuron tracing, pattern analysis and mining, atlas assembling and modeling) to produce this atlas. This picture was used as the cover art of the 2010 Cold Spring Harbor Neuronal Circuit conference, March 10-13, 2010. Particularly, I first produced a 3D neuronal image-pattern atlas by aligning thousands of 3D laser scanning microscopy image-stacks of fruit fly brains (from about 500 GAL4 lines) by using my BrainAligner system (Nature Methods, 2011); then I produced another 3D atlas of stereotyped (spatially invariant) neurite patterns using my Vaa3D-Neuron system (Nature Biotechnology, 2010). |

| The cover picture of Nature Methods (Sept 2009 issue) highlighting the first single-cell resolution 3D digital nuclear atlas of a post-embryonic worm. This atlas documents cell identity of 357 nuclei as well as their statistics of spatial locations, sizes, etc. This atlas is important for single cell level gene expression analysis (with Liu, Long, et al, Cell, 2009) and automatic cell ablation/stimulation. (Read more about this atlas in our Nature Methods 2009 paper). This atlas can also be conveniently viewed/searched in 3D using my Vaa3D software. Indeed, this cover art was generated using Vaa3D as well. |
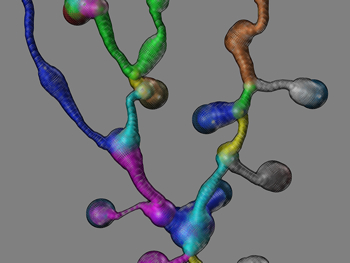
| An artistic painting of a 3D reconstructed and rendered neuron model in adult fruit fly brains. You can download a full-size desktop picture here. This neuron was traced (reconstructed) conveniently using my Vaa3D software (published in Nature Biotechnology, 2010), from 3D confocal images of single neurons produced in Gerry Rubin lab. (Unpublished data. All rights reserved.) |

| The cover art of the Proceedings of the 2009 Bioimage Informatics conference. This picture shows two single neuron reconstructions mapped to an adult fruit fly brain atlas model where various brain compartments are also displayed. The techniques used to trace these neurons and to build the digital atlas are implemented in my Vaa3D software. This picture was produced based on 3D confocal images and painted label-field generated in Julie Simpson, Tzumin Lee, and Gerry Rubin labs. Click here to download a desktop wallpaper picture. (Unpublished data. All rights reserved.) |

The cover art of the Proceedings of the 2008 Conference on Functional Anatomy of the Arthropod Central Complex and Motor System. This picture shows the compartmental atlas of a fruit fly's brain which I produced using the BrainAligner pipeline developed in my lab. The raw image data were produced in Julie Simpson lab. |
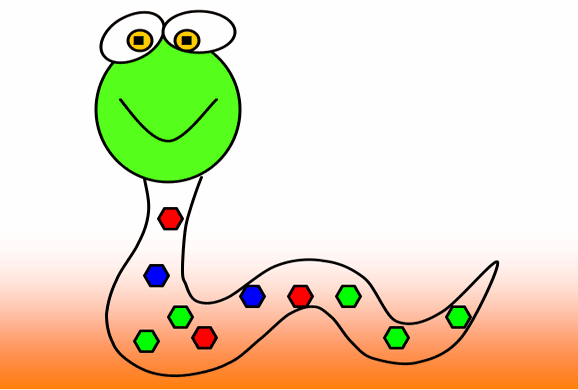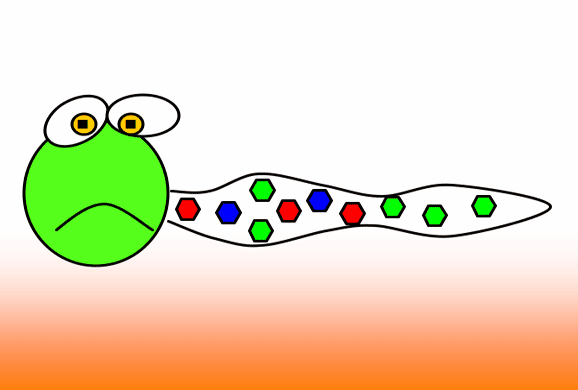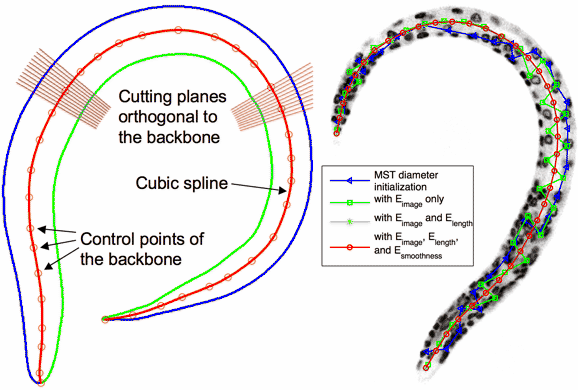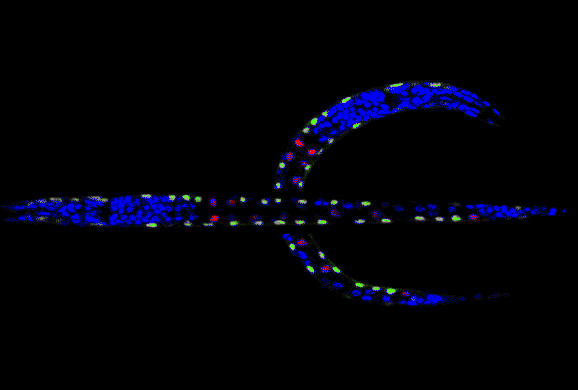
Standardize 3D C. elegans image stacks by straightening the worm body. This technique is one of the critical steps in our project in building the entire digital cell atlas for developing C. elegans. For more information, read a new Bioinformatics paper here. BTW, don't get confused by the above animation: a C. elegans worm does not have eyes! |
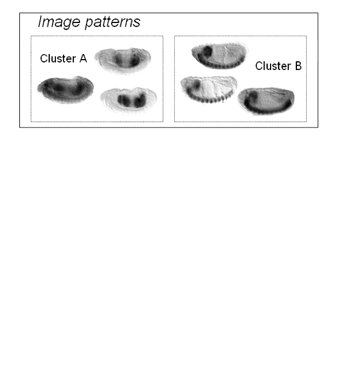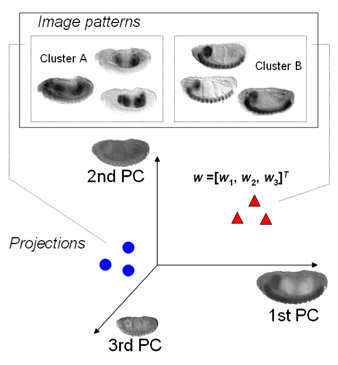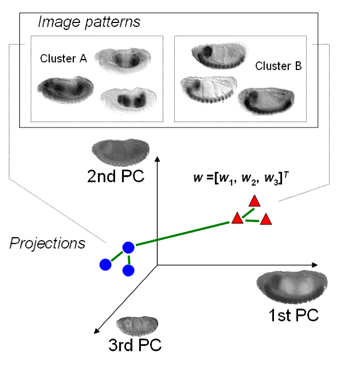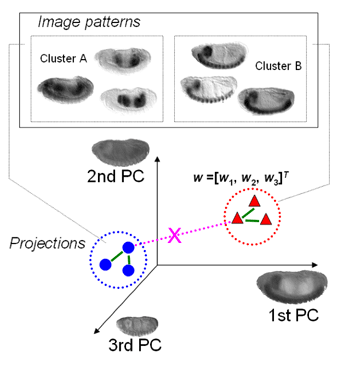
| MSTCUT clustering of fly gene expression patterns in the eigen-embryo space. Read the ISBI2006 paper on the MinMaxPartition MSTCUT method for image clustering and the RECOMB2004 paper on hybrid Gaussian mixture model (GMM) to retrieve genes that have similar expression pattern images. |
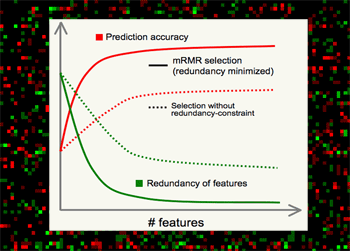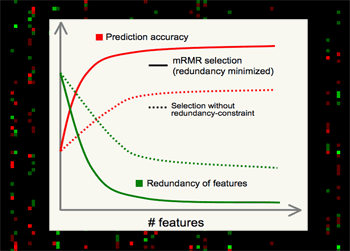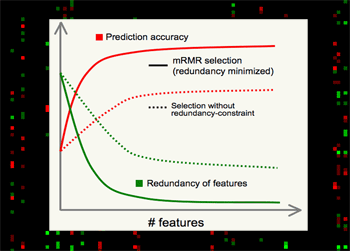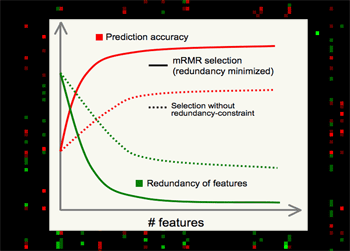
| Minimum Redundancy Maximum Relevance (mRMR) gene selection scheme which is promising in detecting highly discriminant gene markers for cancer classification and is very efficient in optimizing the Max-Dependency/Min-Error feature selection criterion. Read a theory-paper on IEEE Transactions on Pattern Analysis and Machine Intelligence (2005), and a result-paper on Journal of Bioinformatics and Computational Biology (2005). You can run the program here. |
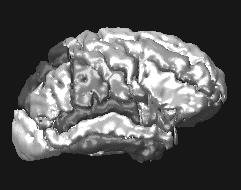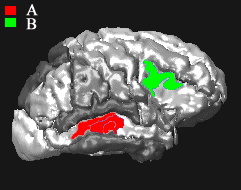
| Automatical detection of brain regions in 3D MR images corresponding to clinical function deficits such as mild cognitive impairment (MCI) and Alzheimer's Disease. Read the Bayesian Morphometry paper in IEEE Transactions on Medical Imaging (2004) and an application study for real MCI data. |
Selected publications[Highly Downloaded | 2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 and Earlier] |
|
DISCLAIMER: These materials are
presented to ensure timely dissemination of scholarly and technical
work. Copyright and all rights therein are retained by authors or by
other copyright holders. All persons copying this information are
expected to adhere to the terms and constraints invoked by each
author's copyright. In most cases, these works may not be reposted
without the explicit permission of the copyright holder. Also note that the software posted below cannot be re-distributed in any form without explicit permission from me. |
Highly Downloaded Papers
Peng, H., Xie, P., Liu, L., .... and Zeng, H.
Liu, L., Yun, Z., Manubens-Gil, L., Chen, H., ..., Peng, H.
Jiang, S., Wang, L., Yun, Z., Chen, H., Liu, L., Yao, J., Peng, H.
Sun, L., Xiong, F., ..., Peng, H.*, Zhao, C.*
Liu, Y., Jiang, S., Li, Y., Zhao, S., Yun, Z., Zhao, Z., ..., Peng, H.
Zhang, L., Huang, L., Yuan, Z., ..., Peng, H.
Qian, P., Manubens-Gil, L., ..., Peng, H.
Manubens-Gil, L., Zhou, Z., Chen, H., Ramanathan, A., Liu, X., ..., Meijering, E., Ascoli, G.A., Peng, H.
Han, X., Guo, S., .... Zhang, L., Peng, H. "Cross-modal coherent registration of whole mouse brains" Lei Qu, Yuanyuan Li, ..., Hanchuan Peng* "Whole-neuron synaptic mapping reveals spatially precise excitatory/inhibitory balance limiting dendritic and somatic spiking" Daniel Maxim Iascone, Yujie Li, Uygar Sumbul, Michael Doron, Hanbo Chen, Valentine Andreu, Finola Goudy, Heike Blockus, Larry F. Abbott, Idan Segev*, Hanchuan Peng*, and Franck Polleux* "TeraVR empowers precise reconstruction of complete 3-D neuronal morphology in the whole brain" Wang, Y., Li, Q., .... and Peng, H. "Automatic tracing of ultra-volumes of neuronal images" Peng, H., Zhou Z., Meijering, E., Zhao, T., Ascoli, G., and Hawrylycz, M. "SmartScope2: simultaneous imaging and reconstruction of neuronal morphology" Long, B., Zhou, Z., Cetin, A., Ting, J., Gwinn, R., Tasic, B., Daigle, T., Lein, E., Zeng, H., Saggau, P., Hawrylycz, M., and Peng, H. "TeraFly: real-time 3D visualization and 3D annotation of terabytes of multidimensional volumetric images" Bria, A., Iannello, G., Onofri, L., and Peng, H. "BigNeuron: large-scale 3D neuron reconstruction from optical microscopy images" Peng, H., Hawrylycz, M., Roskams, J., Hill, S., Spruston, N., Meijering, E., Ascoli, G.A "Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis" Peng, H., Tang, J., Xiao, H., Bria, A., et al., "Extensible visualization and analysis for multidimensional images using Vaa3D" Peng, H., Bria, A., Zhou, Z., Iannello, G., and Long, F.,
Peng, H., Ruan, Z., Long, F., Simpson, J.H., and Myers, E.W. "BrainAligner: 3D registration atlases of Drosophila brains" Peng, H., Chung, P., Long, F., Qu, L., Jenett, A., Seeds, A., Myers, E.W., and Simpson, J.H. "A 3D digital atlas of C. elegans and its application to single-cell analyses" Long, F.*, Peng, H.*, Liu, X., Kim, S. and Myers, E.W. (*equal contribution)
Peng, H., Long, F., and Ding, C. (Run the mRMR "Bioimage informatics: a new area of engineering biology" Peng, H. 2025 Papers
Liu, L., Yun, Z., Manubens-Gil, L., Chen, H., ..., Peng, H.
Jiang, S., Wang, L., Yun, Z., Chen, H., Liu, L., Yao, J., Peng, H.
Sun, L., Xiong, F., ..., Peng, H.*, Zhao, C.* 2024 Papers
Liu, Y., Jiang, S., Li, Y., Zhao, S., Yun, Z., Zhao, Z., ..., Peng, H.
Zhang, L., Huang, L., Yuan, Z., ..., Peng, H.
Qian, P., Manubens-Gil, L., ..., Peng, H.
Xiong, F., Xie, P., ..., Peng, H.
Li, Y., .... , Peng, H., Qu, L.
Li, Y., .... , Peng, H., ..., Ge, W., Zhao, H. 2023 Papers
Manubens-Gil, L., Zhou, Z., Chen, H., Ramanathan, A., Liu, X., ..., Meijering, E., Ascoli, G.A., Peng, H.
Han, X., Guo, S., .... Zhang, L., Peng, H.
Hawrylycz, M., Martone, M., .... , Peng, H., ....
Liu, Y., .... , Peng, H. 2022 Papers"Cross-modal coherent registration of whole mouse brains" Lei Qu, Yuanyuan Li, ..., Hanchuan Peng* 2021 Papers"Morphological diversity of single neurons in molecularly defined cell types" Peng, H., Xie, P., Liu, L., .... and Zeng, H. "Cellular anatomy of the mouse primary motor cortex" Munoz-Castaneda, R., Zingg, B., Matho, K., ..., Peng, H., ... and Dong, H. "Human neocortical expansion involves glutamatergic neuron diversification" Berg, J., Sorensen, S., Ting, J., ..., Peng, H., ... and Lein, E. "A multimodal cell census and atlas of the mammalian primary motor cortex" BRAIN Initiative Cell Census Network (BICCN) "Image enhancement to leverage the 3D morphological reconstruction of single-cell neurons " Guo, S., Zhao, X., .... and Peng, H. 2020 Papers"Whole-neuron synaptic mapping reveals spatially precise excitatory/inhibitory balance limiting dendritic and somatic spiking" Daniel Maxim Iascone, Yujie Li, Uygar Sumbul, Michael Doron, Hanbo Chen, Valentine Andreu, Finola Goudy, Heike Blockus, Larry F. Abbott, Idan Segev*, Hanchuan Peng*, and Franck Polleux* "Binocular encoding in the damselfly pre-motor target tracking system" Jack A. Supple, Daniel Pinto-Benito, Christopher Khoo, ..., Hanchuan Peng, Robert M. Olberg, Paloma T. Gonzalez-Bellido 2019 Papers"TeraVR empowers precise reconstruction of complete 3-D neuronal morphology in the whole brain" Wang, Y., Li, Q., .... and Peng, H. 2017 Papers"Automatic tracing of ultra-volumes of neuronal images" Peng, H., Zhou Z., Meijering, E., Zhao, T., Ascoli, G., and Hawrylycz, M. "SmartScope2: simultaneous imaging and reconstruction of neuronal morphology" Long, B., Zhou, Z., Cetin, A., Ting, J., Gwinn, R., Tasic, B., Daigle, T., Lein, E., Zeng, H., Saggau, P., Hawrylycz, M., and Peng, H. "Fast assembling of neuron fragments in serial 3D sections" Chen, H., Iascone, D.M., da Costa, N., Lein, E., Liu, T., and Peng, H., "Ensemble neuron tracer for 3D neuron reconstruction" Wang, C.W., Lee, Y., Pradana, H., Zhou, Z., and Peng, H.. "3D neuron tip detection in volumetric microscopy images using an adaptive ray-shooting model" Liu, M., Gong, R., Chen, W., and Peng, H. 2016 Papers"TeraFly: real-time 3D visualization and 3D annotation of terabytes of multidimensional volumetric images" Bria, A., Iannello, G., Onofri, L., and Peng, H. "Imagining the future of bioimage analysis" Meijering, E., Carpenter, A., Peng, H., Hamprecht, F., and Olivo-Marin, J.C. "To the cloud! A grassroots proposal to accelerate brain science discovery" Neuro Cloud Consortium "Inferring cortical function in the mouse visual system through large-scale systems neuroscience" Hawrylycz, M., Anastassiou, C., ..., Peng, H., ... "Rivulet: 3D Neuron Morphology Tracing with Iterative Back-Tracking" Liu, S.Q., Zhang, D., Li, S.D., Feng, D., Peng, H., and Cai, W.D. "Bioimage informatics for big data" Peng, H., Zhou, J., Zhou, Z., Bria, A., Li, Y., Kleissas, D.M., Drenkow, N.G., Long, B., Liu, X., and Chen, H. "TReMAP: automatic 3d neuron reconstruction based on tracing, reverse mapping and assembling of 2d projections" Zhou, Z, Liu, X., Long, B., and and Peng, H. 2015 Papers"3D Image-guided automatic pipette positioning for single cell experiments in vivo" Long, B., Li, L., Knoblich, U., Zeng, H., and Peng, H. "BigNeuron: large-scale 3D neuron reconstruction from optical microscopy images" Peng, H., Hawrylycz, M., Roskams, J., Hill, S., Spruston, N., Meijering, E., Ascoli, G.A "From DIADEM to BigNeuron" Peng, H., Meijering, E., Ascoli, G., "Blastneuron for automated comparison, retrieval and clustering of 3d neuron morphologies" Wan, Y., Long, F., Qu, L., Xiao, H., Hawrylycz, M., Myers, E., Peng, H., "SmartTracing: self-learning based neuron reconstruction" Chen, H., Xiao, H., Liu, T., and Peng, H. "3-D registration of biological images and models: registration of microscopic images and its uses in segmentation and annotation" Qu, L., Long, F., Peng, H., "Interactive exemplar-based segmentation toolkit for biomedical image analysis" Li, X., Zhou, Z., Keller, P., Zeng, H., Liu, T., and Peng, H., 2014 Papers"Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis" Peng, H., Tang, J., Xiao, H., Bria, A., et al., "Extensible visualization and analysis for multidimensional images using Vaa3D" Peng, H., Bria, A., Zhou, Z., Iannello, G., and Long, F., "Adaptive image enhancement for tracing 3d morphologies of neurons and brain vasculatures" Zhou, Z., Sorensen, S., Zeng, H., Hawrylycz, M., and Peng, H., "LittleQuickWarp: an ultrafast image warping tool" Qu, L., Peng, H., "A mesoscale connectome of the mouse brain" Oh, S.W., Harris, J.A., Ng, L., ..., Peng, H., ..., Zeng, H., "Constructing 5D developing gene expression patterns without live animal imaging" Peng, H., Myers, E.W., "Atlas-builder software and the eNeuro atlas: resources for developmental biology and neuroscience" Heckscher, E.S., Long, F., ..., Peng, H., Myers, E.W., Doe, C., "Wiring economy can account for cell body placement across species and brain areas" Rivera-Alba, M., Peng, H., de Polavieja G.G., and Chklovskii, D.B., 2013 Papers"Automated image computing reshapes computational neuroscience" Peng, H., Roysam, B., and Ascoli, G., "APP2: automatic tracing of 3D neuron morphology based on hierarchical pruning of gray-weighted image distance-trees" Xiao, H., and Peng, H., "BIOCAT: a pattern recognition platform for customizable biological image classification and annotation" Zhou, J., Lamichhane, S., Sterne, G., Ye, B., and Peng, H., "Micron-scale resolution optical tomography of entire mouse brains with confocal light sheet microscopy" Silvestri, L., Bria, A., Costantini, I., Sacconi, L., Peng, H., Iannello, G., and Pavone, F.S., "Eight pairs of descending visual neurons in the dragonfly give wing motor centers accurate population vector of prey direction" Gonzalez-Bellido, P., Peng, H., Yang, J., Georgopoulos, A.P., and Olberg, R.M., "A distance-field based automatic neuron tracing method" Yang, J., Gonzalez-Bellido, P., and Peng, H. "Automated cellular annotation for high resolution images of adult C. elegans" Aerni, SJ., Liu, X., Do, CB, Gross, SS, Nguyen, A., Guo, SD, Long, F., Peng, H., Kim, SS, and Batzoglou, S., "Clonal development and organization of the adult Drosophila central brain" Yu, H.H., Awasaki, T., Schroeder, M., Long, F., Yang, J.S., He, Y., Ding, P. Kao J., W. G., Peng, H., Myers, G., and Lee, T., 2012 Papers"Visualization and analysis of 3D microscopic images" Long, F., Zhou, J., and Peng, H. "A GAL4-driver line resource for Drosophila neurobiology" Jenett, A., Rubin, G., ..., Peng, H., ..., and Zugates, C. "Biological imaging software tools" Eliceiri, K.W., Berthold, M.R., Goldberg, I.G., Ibanez, L., Manjunath, B.S., Martone, M.E., Murphy, R.F., Peng, H., Plant, A.L., Roysam, B., Stuurmann, N., Swedlow, J.R., Tomancak, P., and Carpenter, A.E. "mGRASP enables mapping mammalian synaptic connectivity with light microscopy" Kim, J., Zhao, T., Petralia, R.S., Yu, Y., Peng, H., Myers, E.W., and Magee, J.C., "Bioimage informatics: a new category in Bioinformatics" Peng, H., Bateman, A., Valencia, A., and Wren, J.D. 2011 Papers"BrainAligner: 3D registration atlases of Drosophila brains" Peng, H., Chung, P., Long, F., Qu, L., Jenett, A., Seeds, A., Myers, E.W., and Simpson, J.H. "Simultaneous recognition and segmentation of cells: application in C. elegans" Qu, L., Long, F., Liu, X., Kim, S., Myers, E.W., and Peng, H. "Automatic 3D neuron tracing using all-path pruning" Peng, H., Long, F., and Myers, E.W. "Proof-editing is the bottleneck of 3D neuron reconstruction: the problem and solutions" Peng, H., Long, F., Zhao, T., and Myers, E.W. "Automated high speed stitching of large 3D microscopic images" Yu, Y., and Peng, H. 2010 and Earlier"V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets" Peng, H., Ruan, Z., Long, F., Simpson, J.H., and Myers, E.W. "Automatic reconstruction of 3D neuron structures using a graph-augmented deformable model" Peng, H., Ruan, Z., Atasoy, D., and Sternson, S. "A principal skeleton algorithm for standardizing confocal images of fruit fly nervous systems " Qu, L. and Peng, H. "Segmentation of center brains and optic lobes in 3D confocal images of adult fruit fly brains" Lam, S.C.B., Ruan, Z., Zhao, T., Long, F., Jenett, A., Simpson, J.H., Myers, E.W., and Peng, H. "A 3D digital atlas of C. elegans and its application to single-cell analyses" Long, F.*, Peng, H.*, Liu, X., Kim, S., and Myers, E.W. (*equal contribution) "Analysis of cell fate from single-cell gene expression profiles in C. elegans" Liu, X., Long, F., Peng, H., Aerni, S.J., Jiang, M., Sanchez-Blanco, A., Murray, J.I., Preston, E., Mericle, B., Batzoglou, S., Myers, E.W., and Kim, S. "VANO: a volume-object image annotation system" Peng, H., Long, F., and Myers, E.W. "Bioimage informatics: a new area of engineering biology" Peng, H. "Automatic recognition of cells (ARC) for 3D images of C. elegans" Long, F., Peng, H., Liu, X., Kim, S., and Myers, E.W. "Straightening C. elegans images" Peng, H., Long, F., Liu, X., Kim, S., and Myers, E.W. "Data driven decomposition for multi-class classification" Zhou, J., Peng, H., and Suen, C.Y. "Automatic image analysis for gene expression patterns of fly embryos" Peng, H., Long, F., Zhou, J., Leung, G., Eisen, M.B., and Myers, E.W. "Automatic recognition and annotation of gene expression patterns of fly embryos" Zhou, J.*, and Peng. H.* (*equal contribution) "Phenotype clustering of breast epithelial cells in confocal images based on nuclear protein distribution analysis" Long, F., Peng, H., Sudar, D., Lelievre, S., and Knowles, D. "Transitive closure and metric inequality of weighted graphs - detecting protein interaction modules using cliques," Ding, C., He, X., Xiong, H., Peng, H., and Holbrook, S.R. "Clustering gene expression patterns of fly embryos," Peng, H., Long, F., Eisen, M.B., and Myers, E.W.
Peng, H., Long, F., and Ding, C. (Run the mRMR "Minimum redundancy maximum relevance feature selection," Peng, H., Ding, C., and Long, F. "Minimum redundancy feature selection from microarray gene expression data," Ding, C., and Peng, H. (A conference version with a different set of results, but the same title, also appeared on: "Comparing in situ mRNA expression patterns of Drosophila embryos," Peng, H., and Myers, E.W. "A Bayesian morphometry algorithm," Herskovits, E., Peng, H., and Davatzikos, C. (This is an extended journal version of the following earlier ISBI paper.) "Bayesian clustering methods for morphological analysis of MR images," Peng, H., Herskovits, E.H., and Davatzikos, C., An electronic demo were also given at 2002 NIH Human Brain Project Annual Meeting, A study using real BLSA Mild Cognitive Impairment data is the following report: "Bayesian analysis of morphological changes associated with mild cognitive impairment: a cross-sectional study," Peng, H., Shen, D., Davatzikos, C., Resnick, S., and Herskovits, E.H. "Structural search and stability enhancement of Bayesian networks," Peng, H., and Ding, C. "Document image recognition based on template matching of component block projections," Peng, H., Long, F., and Chi, Z. |
 "Morphological diversity of single neurons in molecularly defined cell types"
"Morphological diversity of single neurons in molecularly defined cell types" "Connectivity of single neurons classifies cell subtypes in mouse brains"
"Connectivity of single neurons classifies cell subtypes in mouse brains"