-
•Vaa3D: 3D/4D/5D image/surface renderer, 3D image analysis & meta-data management, and more (highly downloaded)
-
•Vaa3D Matlab IO toolbox: some Matlab routines to read/write V3D data files for further analysis
-
•CellExplorer: 3D C. elegans atlas building & application tools
-
•VANO: 3D image annotator
-
•BDB-: deformable curve in n-dim
-
•mRMR: feature selector (highly accessed)
-
•Mutual information Matlab toolbox: (highly downloaded)

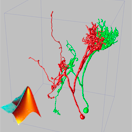
Vaa3D (The Swiss army knife for 3D/4D/5D image visualization & analysis) (Current version 2.7)
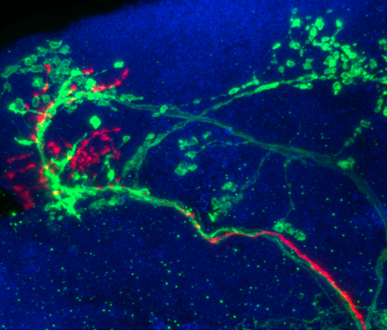
A handy tool for visualizing and analyzing large 3D image stacks and various surface data. Not only a fast 3D renderer, but also a container of powerful 3D image analysis (cell segmentation, neuron tracing, brain registration, annotation, etc) and data management modules. Customary plugins can be developed by users. With the versatile 3D viz-analysis features and quantitative statistical measurements, Vaa3D provides an excellent desktop solution for various bioimage informatics applications, and also a platform to develop new 3D image analysis algorithms which can be deployed for high-throughput processing. Vaa3D streamlines the viz-analysis workflow, - the whole is greater than the sum of its parts!

Vaa3D Matlab IO toolbox [ Download it at Vaa3D website ]
Read/write basic Vaa3D data files (image files .RAW, .TIFF, marker file .marker, point cloud file .apo, neuron file .swc).
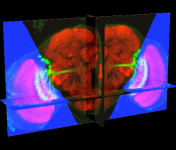
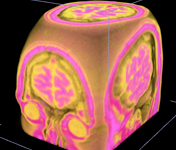
[ Bioinformatics-2009 Paper | BII09 software-highlight slides ]
A well-coordinated way to annotate hundreds or thousands of 3D image objects. Combine 3D views of images and spread sheet neatly. Just easy to manage 3D segmented image objects, or let you incorporate your segmentation priors, or let you edit your segmentation results!
This system is used in building digital atlases of C. elegans, fruit fly embryos at single cell (nucleus) resolution, and the compartment-level of digital map(s) of adult fruit fly brains.

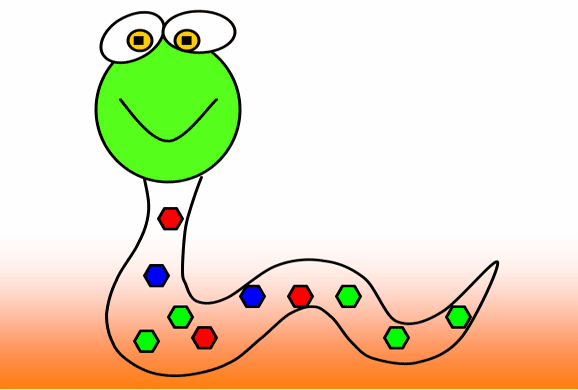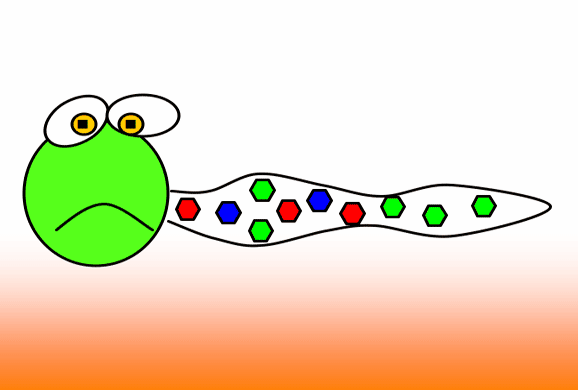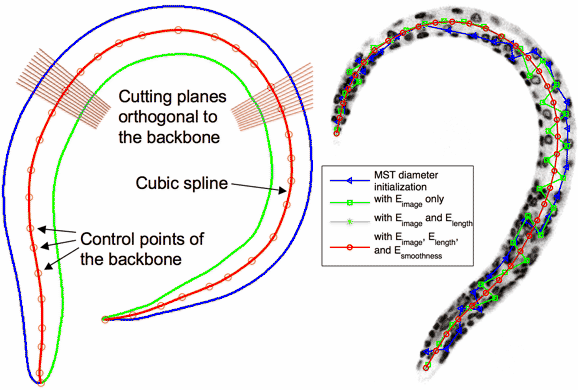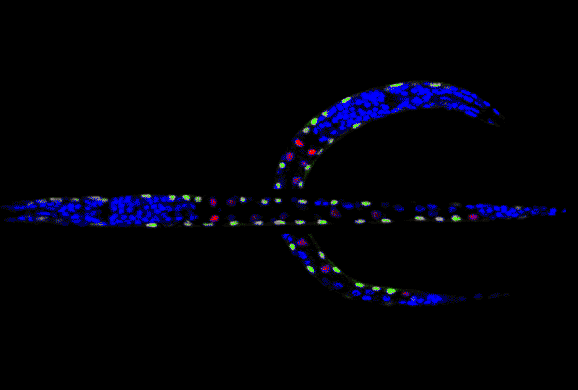
BDB- [ Bioinformatics-2008 Paper ]
Backbone Detection without Boundary (BDB-) method can find the curved smooth principal axis that passes through the "middle" of an elongated image object, even the object has no boundary. This method can be used in N-dimensional space, and has been tested for real applications like straightening the curved C. elegans body.


mRMR (highly accessed!)
[ TPAMI-2005 Paper | JBCB-2005/CSB'03 Paper ]
minimum-Redundancy-Maximum-Relevance (mRMR) feature selection is among the most powerful methods to select a subset of features from a big pool, or just do dimension reduction. This method has been well cited and used for many different applications.


Mutual Information Toolbox (Highly downloaded!)
Simple Matlab toolbox to compute mutual information. This toolbox is also needed if you run the Matlab version of my mRMR feature selector.


[ Nature Methods 2009 Paper | Cell 2009 paper ]
The pipeline to produce the first 3D digital atlas of a post-embryonic worm at the single cell resolution, and to automatically predict cell identity of a new image. Can be re-used for other systems (e.g. fruitfly).
This software package was mostly developed by Fuhui Long, - but I shamelessly host it at my website, :-)).
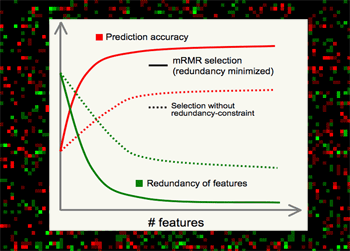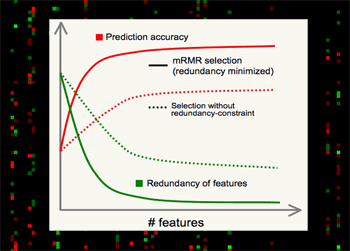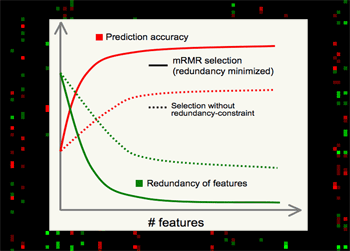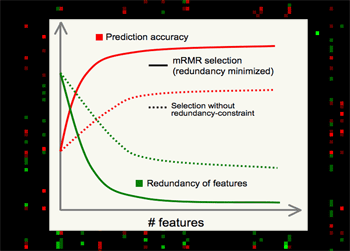
mRMR is valuable for several major reasons.
-
•Theoretically, it's shown that high-dimensional combinatorial Max-Dependency search can be efficiently reduced as solving a series of 2-dimensional problems of mRMR, which is much faster, and in practice more robust.
-
•Outstanding performance in selecting features, - always much better than relevance-only selection schemes. mRMR features normally yield excellent classification accuracy on different classifiers (e.g. SVM, LDA, Naive Bayes, KNN, neural networks, logistic regression, etc).
-
•Can be easily combined with other feature selectors of different types, such as wrappers.
-
•Simple!